Publications
26. Bolshakov, D.T., Weix, E.W.Z, Galateo, T.M., Rajasekaran, R., Coyle, S.M. Noise-guided tuning of synthetic protein waves in living cells. ACS Synthetic Biology, (2025). doi: https://doi.org/10.1021/acssynbio.5c00599
Link to original BioRxiv preprint
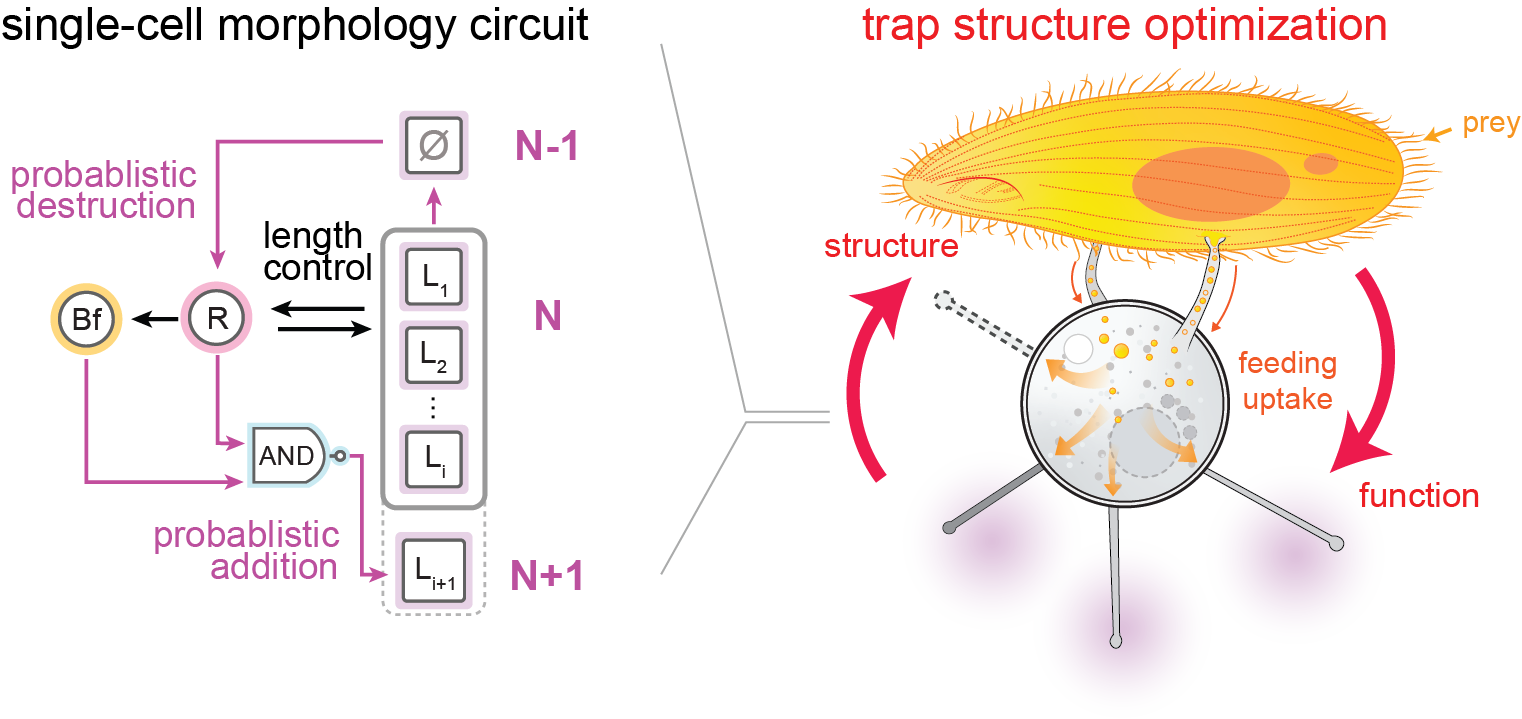
25. Xu, Z., Mazurkiewicz,L.E., Olivetta, M., Morrissey, M., Dudin, O., Weeks, A.M., Coyle, S.M. A self-organizing single-cell morphology circuit optimizes Podophrya collini predatory trap structure. bioRixv, preprint (2025).
suctorian-viewer repository and data
25. Chandrasekharan, N., Lei, X., Honts, J. and Bhamla, S., Coyle, S.M. Decoding ultrasensitive self-assembly of the calcium-regulated Tetrahymena cytoskeletal protein Tcb2 using optical actuation. Journal of Biological Chemistry. In press. 110824 October (2025)
Original bioRxiv, preprint (2025).
23. Lei, X., Floyd, C., Ferrer, L.C., Chakrabortty, T., Chandrasekharan, N., Dinner, A., Coyle, S.M., Honts, J. and Bhamla, S., Light-induced reversible assembly and actuation in ultrafast Ca2+-driven chemomechanical protein networks. bioRxiv, preprint (2025).
22. Rajasekaran, R. Galateo, T.M., Xu, Z., Bolshakov, D.T., Weix, E.W.Z., Coyle, S.M. Genetically encoded protein oscillators for FM streaming of single-cell data. bioRxiv preprint (2025).
21. Xu,Z. Chang, C.C., Coyle, S.M. Synthetic forms most beautiful: engineering insights into self-organization. Physiology (2025). https://doi.org/10.1152/physiol.00064.2024
20. Chang, C.C., Coyle, S.M. Regulatable assembly of synthetic microtubule architectures using engineered MAP-IDR condensates. Journal of Biological Chemistry, Special Issue: Cell and Developmental Biology. 300 (8), 107544 (2024). PDF.
Link to original bioRxiv preprint (2023).
19. Rajasekaran, R., Chang, C.C., Weix, E.W.Z., Galateo, T.M., Coyle, S.M. A programmable reaction-diffusion system for spatiotemporal cell signaling circuit design. Cell 187, 345–359 (2024).
Supplemental Circuit Design Manual
Original bioRxiv Preprint:
Main Text.
Supplemental Materials and Methods.
Supplemental Movies Playlist.
18. Ahn, C.J., Coyle, S.M. Comparative profiling of cellular gait on adhesive micropatterns defines statistical patterns of activity that underlie native and cancerous cell dynamics. bioRxiv Preprint (2023).
17. Coyle, S. M., Ciliate behavior: blueprints for dynamic cell biology and microscale robotics. Molecular Biology of the Cell (22), 2415-2420 (2020).
16. Coyle, S. M., Flaum, E., Li, H., Krishnamurthy, C., Prakash, M. Coupled active systems encode an emergent hunting behavior in the unicellular predator Lacrymaria olor. Current Bio\logy 29 (22), 3838-3850 (2019).
Featured Current Biology dispatch: Ciliate Biology: The Graceful Hunt of a Shape-Shifting Predator. (Kirsty Wan)
15. Coyle, S.M. Reverse engineering GTPase programming languages with reconstituted signaling networks. Small GTPases 7 (3), 168-172 (2016).
14. Morsut, L., Roybal, K.T., Xiong, X., Gordley, R.M., Coyle, S.M., Thomson, M., Lim, W.A. Engineering customized cell sensing and response behaviors using synthetic notch receptors. Cell 164, 780-791 (2016).
13. Coyle, S.M., Lim, W.A. Mapping the functional versatility and fragility of Ras GTPase signaling circuits through in vitro network reconstitution. eLife, 5:e12345 (2016).
• Faculty of 1000 recommended selection.
• Striking image highlighted on eLife front page.
12. Dumesic, P.A., Homer, C.M., Moresco, J.J., Pack, L.R., Coyle, S.M., Strahl, B.D., Fujimori, D.G., Yates, J.R., Madhani, H.D. Product binding directs genomic specificity of a yeast Polycomb repressive complex. Cell 160, 204-216 (2015).
11. Van Anken, E., Pincus, D., Coyle, S.M., Aragon, T., Osman, C., Lari, F., Puerta, S.G., Korennykh, A.V., Walter, P. Specificity in endoplasmic reticulum stress signaling entails a step-wise engagement of HAC1 mRNA to Ire1 clusters. eLife, 3:e05031 (2014).
10. Coyle S.M., Flores J., Lim W.A. Exploitation of latent allostery enables the evolution of new modes of MAP kinase regulation. Cell 154, 875-887 (2013).
9. Kroll E., Coyle S., Dunn B., Koniges G., Aragon A., Edwards J., Rosenzweig F. Starvation-associated genome restructuring can lead to reproductive isolation in yeast. PLoS ONE 8, e66414 (2013).
8. Zalatan J.G.*, Coyle S.M.*, Rajan S., Sidhu S.S., Lim W.A. Conformational control of the Ste5 scaffold protein insulates against MAP kinase misactivation. Science 337, 1218-1222 (2012).
• Perspective in Science by Roger Davis.
• Editor’s Choice, Science Signaling.
7. Rougemaille M., Braun S.*, Coyle S.*, Dumesic P.A.*, Garcia J.F.*, Isaac R.S.*, Libri D., Narlikar G.J., Madhani H.D. Ers1 links HP1 to RNAi. Proc. Natl. Acad. Sci. 109, 11258-11263 (2012).
6. Jinek M., Coyle S.M, Doudna J.A. Coupled 5’ nucleotide recognition and processivity in Xrn1-mediated mRNA decay. Mol. Cell 41, 600-608 (2011).
5. Weeks A.M., Coyle S.M., Jinek M., Doudna J.A., Chang M.C.Y. Structural and biochemical studies of a fluoroacetyl-CoA-specific thioesterase reveal a molecular basis for fluorine selectivity. Biochemistry 49, 9269-9279 (2010).
4. Jinek M., Fabian R.M., Coyle S.M., Sonenberg N., Doudna J.A. Structural and biochemical analysis of the human GW182-PABC interaction in microRNA-mediated silencing. Nat. Struct. Mol. Biol.17, 238-240 (2010).
3. Wiedenheft B., Zhou K., Jinek M., Coyle S.M., Ma W., Doudna J.A. Structural basis for DNase activity of a conserved protein implicated in CRISPR-mediated genome defense. Structure 17, 904-912 (2009).
2. Coyle S.M., Gilbert W.V., Doudna J.A. Direct link between RACK1 function and localization at the ribosome in vivo. Mol. Cell. Biol. 29, 1626-1634 (2009).
1. Coyle S., Kroll E. Starvation induces genomic rearrangements and starvation-resilient phenotypes in yeast. Mol. Biol. and Evol. 25, 310-318 (2008).